During this visit, you’ll learn about:
Developmental and gender related psychosocial differences in pain sensitivity, vulnerability, pain experiences, expressions, and treatment disparity and efficacy.
How the sociocultural background of the patient, as well as the social setting, influence perception, communication, tolerance, response, coping strategies, and treatment.
How genetic and epigenetic factors of pain susceptibility are differentiated
The multigenic nature of pain, and examples of contributions to pain sensitivity and chronic pain will be identified
Developmental and gender related biological differences in pain sensitivity, vulnerability, pain experiences, expressions, and treatment disparity and efficacy
If you need to review of the basics of human genetics, explore these NIH and NLM links:
Fact Sheets on Science, Research, Ethics and the Institute (https://www.genome.gov/about-genomics/fact-sheets)
Genetics Home Reference Glossaries and Medical Terms (http://ghr.nlm.nih.gov/glossary=genetics)
Psychosocial Factors
Biopsychosocial Model of Pain
Different people express their pain differently. Cultural norms weigh heavily on how people express pain. If a male has internalized the norm of the “tough guy”, then he may under-report his pain and under utilize pain management strategies, whereas females may be more expressive with their healthcare providers and communicate more about their pain due to this being more socially acceptable.
There are norms for EVERY GROUP IMAGINABLE, and it is important to appreciate these distinctions and expectations for how much pain is reported, how its impact on daily function is assessed, how it is described, and how the patient’s expectations for pain management may be different as a result.
This could greatly shape how open patients are about their pain and also how open they are to management. The role of the healthcare team is to help the patients achieve the best possible outcome but this requires understanding and unbiased application of the biopsychosocial model of pain for the most complete assessment of the patient’s pain status and symptom burden that may impact pain management efficacy.
Biological factors, specifically the genetic and physiological mechanisms of pain, will be taught throughout the course, but these factors cannot be considered in isolation from the sociocultural and psychological factors.
Biological Factors include tissue damage, genetic factors, physiological mechanisms of pain
However, social aspects set the framework for how we react to pain. They determine what is acceptable as well as what is taboo in how we deal with pain.
Sociocultural Factors include ethnicity, family history and cultural factors.
Culture sets boundaries for how we experience and describe our pain. Several research reports highlight the fact that people from different cultural heritages respond somewhat differently to painful stimuli.
Psychological Factors include negative mood (anger, anxiety, depression), coping strategies and social learning.
Situational Factors that Modify Pain Perception
When assessing and treating pain, it is important to appreciate that many factors can play a role in coping with pain. Developmental factors have an impact on:
- Perception of pain
- Understanding of the meaning of pain (e.g. cause, treatment)
- Expression of pain (e.g. verbal, behavioral)
- Suitability of different assessment and treatment options
Regardless of its cause, the experience of pain is always influenced by a complex interaction of physical, cognitive, emotional, social and behavioral factors.
Family and members of the broader treatment team play an important role in the assessment and treatment process. It is essential to have a shared understanding of appropriate expectations for pain management and help to support the patient’s use of effective coping strategies and to promote function.
Questioning whether the patient’s pain is “real”, or of the reported severity or intensity, is not helpful. Treatment works best when all treatment team members communicate an appreciation of the biopsychosocial model of pain as early as possible in the diagnostic work up.
Assessment
Let’s look at some additional information to allow a more complete understanding of the impact of the patient’s pain problem and factors that may be serving to maintain or exacerbate it.
To assess the psychosocial dimensions of pain, it is helpful to learn:
- the context and consequences of the pain
- the patient’s patterns of coping
- the presence of stressors
- the presence of anxiety, withdrawal, attention and learning problems
Behavioral Factors
Consider the role of family members’ behaviors (e.g. modeling, reinforcement) and factors (e.g. family functioning, cohesion).
Complex interrelationships exist among family members‘ beliefs and worries about a loved one’s pain, pain promoting behavior, disability, family functioning, and stress.
These relationships are important to consider as they may have implications for treatment decisions.
Emotional Factors
The experience of pain does not solely rely on noxious stimuli, but an interplay of variables which modulate the pain experience, including memory, mood, environment, attention and expectation.
Evidence for pain modulatory mechanisms was first documented by Henry Beecher, a medic serving in the U.S. Army during World War II. While treating wounded American soldiers, Beecher ran out of pain-killing morphine. Desperate, he decided to continue telling the soldiers that he was giving them morphine, although he was actually infusing them with a saline solution. Amazingly, 40 percent of the soldiers reported that the saline treatment eased their pain and did not require analgesics for pain. He reported these men were alert and responsive though the injuries included compound fractures and penetrating wounds, concluding that "strong emotions" can block pain. Ultimately, this means pain experienced by different individuals from the same noxious stimuli can vary considerably.
Underpinning the descending pain modulatory system is the endogenous opioid system. This system may be activated by a variety of cognitively-trigged states. The endogenous opioid system causes inhibition of substance P from peripheral noxious mechanical stimulation via release of noradrenaline from the dorsalateral PAG (dPAG) and thermal nociceptive stimuli via the release of serotonin from the ventrolateral PAG (vPAG).

*Courtesy of the National Library of Medicine and the United States Army.
Apply Your Knowledge
Now, consider your patient with jaw clicking without and with pain, and frequent headaches. Ask yourself the following questions:
- Is this patient female or male?
- Of African-American, Irish, Italian, Egyptian, Somoan, Japanese, Chinese, Native American, other, or mixed heritage?
- How old is the patient?
- Has the patient had previous experience with pain?
- Who are the patient’s pain models?
- What stresses may impact the patient’s pain perception?
- What support system does the patient have?
- How is the patient coping with pain?
- How do these biological, psychological and sociocultural factors impact the patient's pain, care planning and your treatment decisions?
Patient Information
- Your patient is actually a 15 year old female named Melanie who reports jaw clicking without and with pain, and frequent headaches for 3 months.
- She lives with her parents and two sisters, one a senior in the same high school, the other a fifth grader. The family has a dog, a cat and a bird; the dog belongs to your patient.
- No one else in the household has been ill in the past six months.
- Your patient is a good student (A’s and B’s, some honors courses). She is active on her high school debate team and student government. She has a positive peer group and several best friends.
- Menarche began at 12 years of age. She has had regular monthly menses for about a year with mild discomfort, relieved with ibuprofen.
- She denies dating and any sexual activity. She also denies use of tobacco, alcohol, or drugs.
Pain Threshold, Sensitivity and Tolerance
Nurse Allele, BSN, RN asks:
On a scale of 0 to 10, with 0 being no pain and 10 being:
- the worst pain you have ever experienced
- the worst pain you can imagine
- the most pain you can tolerate
- At what number would you call this experience pain?
- How much does your jaw hurt?
Pain Variation
Individuals experience variation in pain because of variation in the painful stimuli, such as the type of tissue trauma, severity of trauma, or location. This cause and effect model simplifies pain as a yes/no phenomenon, but pain is actually a continuous-variable, and similar stimuli results in great inter- and intra- individual variability (i.e. the same strength stimulus is not ranked the same by everyone on that 0-10 scale).
This variability can be described not only in terms of intensity (how much pain, how painful), but also duration (how long has it been painful), quality (what does the pain feel like), and its function (how does the pain make you feel or impact your life).
- Type
- Severity
- Location
- Intensity
- Duration
- Quality
- Function
There is a normal distribution of a wide range of pain sensitivity/ susceptibility even in so-called “healthy normals”.
This bar graph illustrates the distribution of a summary measure of pain from a sample of a normal population.
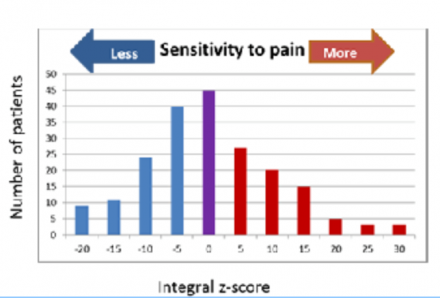
This summary measure of pain sensitivity was derived from sixteen individual pain measures (pain modality), each standardized to unit normal deviates (z-scores) with a mean of 0 and standard deviation of 1.
Individuals represented at the extreme left-side of the figure are resistant to pain-evoking procedures, whereas individuals represented at the extreme right-side of the figure are most sensitive to pain-evoking procedures.
This is why comparable stimuli in different people may produce different intensities of pain experiences, and why patients may sleep or be distracted from pain even when experiencing a high level of pain. This type of continuous variation in normal populations suggests that pain is a complex phenotype influenced by genetic and environmental factors.
Pain Threshold, Sensitivity and Tolerance
The definition of the scale anchor has meaning for the patient.
Threshold
At what number does the patient even call this experience, this sensation, this phenomenon, pain? At a 1, a 2, a 4 ½?
Sensitivity
What is the worst pain you have ever experienced – a stubbed toe, a broken bone, childbirth, sunburn, a shot, a kidney stone? What is the worst pain you can imagine – an elephant standing on your head? a hot poker in the eye?
Tolerance
What is the most pain you can tolerate? an ache, a 3 on a 0 to 10 scale, or perhaps an 8 on a 0 to 10 scale, or you may even pride yourself as having a high tolerance to pain based on athletic performance or your ability to function at work or school despite pain.
How Can Polymorphisms Convey Susceptibility to Pain?
The processes involved in pain, for example ion channel activation, neurotransmitter release, etc., are set biologically and don't vary much. However, variation within the genes that encode for each of the molecules involved in those processes can have SNPs that alter the structure and function of the end products created. This variation then results in small, more nuanced differences in those hardwired processes that translate into differences in pain sensitivity.
In this way, the genetic variation acts as a way of modulating the existing processes involved in pain and can result in differences between people determined by their specific combination of SNPs.
The signaling molecules (cytokines, neurotransmitters, etc.), receptors, channels, etc. that provide the experience of pain (explained in the Neurophysiology Visit) are proteins expressed from genetic code.
Genetic code (https://ghr.nlm.nih.gov/resources#medicalterminology) is set at conception and is unchanged throughout your life, so it is static.
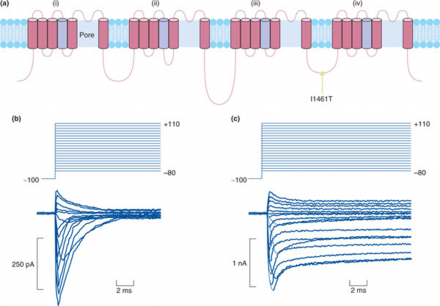
*image: Segerdahl et al. (2012) presented in Crow et al. (2013). Genomic Medicine, 5:12
The “genes” are the same between individuals; for example, all have a SCN9A, pictured here, that encodes the voltage gated sodium channel Nav1.7. However, there are different versions, or alleles, for each gene that occur within populations, and these little variations, or non-synonymous SNPs, may alter pain phenotype. Sometimes these are rare, and may be referred to as a mutation only if they are extremely rare, but a mutation is just a SNP that doesn't occur greater than 1% of the time in the general population.
Single Nucleotide Polymorphisms can confer increased risk for pain by causing changes to the resulting protein structure, which can change protein function. This means that the protein (i.e., the neurotransmitter, enzyme, receptor, etc.) is structurally different, and this can affect structure and/or function, resulting in clear differences in the way the protein works.
In this case, a SNP causing a change from an isoleucine to threonine residue (indicated by the substituted amino acid and the location as I1461T) in the loop domain leads to loss of channel inactivation, which is responsible for inherited paroxysmal pain disorder.
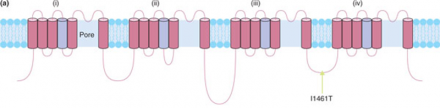
Diagram (b) shows the action potential firing from a human embryonic kidney cell with a "normal" isoleucine containing Nav1.7 channel.
You can see that the cell membrane potential changes quickly after the initial stimulus and then resolves back to the cellular resting potential quickly with a stereotyped shape.
Whereas diagram (c) shows the same types of cells but containing the SNP with the rare allele resulting in a threonine substitution in one of the intracellular loops (I1461T).
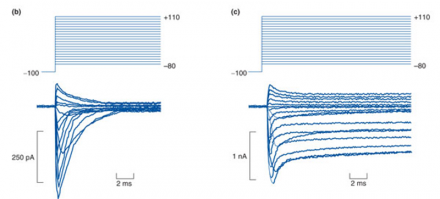
These cells have abnormally long action potentials that do not resolve back to resting potential. If you think of these action potentials as "pain signals" or "pain messages", then you can see that the normal cells send a "pain signal" that is short and resolves; this is normal.
The cells with the SNP of interest would have a VERY prolonged "pain signal" and do not show the resolution back to rest. So, this would result in more pain messages coming in and being processed by the brain, even if the number and intensity of stimuli was identical to that shown in diagram b.
For other SNPs, we don’t know exactly why those particular SNPs are associated with pain risk. If a SNP is in a non-coding region of the genome, or if it is synonymous, meaning that the ending amino acid sequence that encodes the protein end-product of gene expression is unaltered, then it is not always clear why that SNP is associated with different outcomes.
These relationships, however, can be excellent for generating hypotheses related to pain processes/modulation and, as a result, function of the genes may be better understood.
Key Genes and Function in Pain Processing
Pain is multigenic in addition to the environmental factors that contribute to pain sensitivity. We have identified a number of genes that explain some of these individual differences in pain sensitivity (both in normals and in patient populations).
This figure shows genes (by chromosomal location) where variations (SNPs) have been implicated in pain sensitivity. Each color dot represents a different type of pain.
You can see that there are genes that appear to influence pain in multiple modalities (nonspecific) and those that have only been associated with a single pain time (specific). It is important to note that we are not talking about mutations here. We’re not talking about “alleles” that are highly rare. We’re talking about variation that is common and which can contribute to variation in the outcome of interest (in this case, pain).
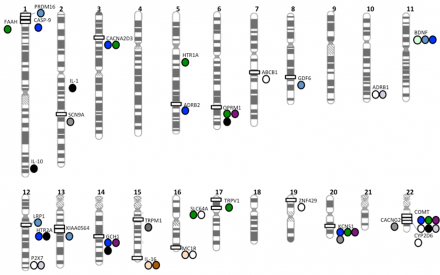
*Image courtesy of Young et al. (2012). Journal of Medical Genetics, 49(1), 1-9.
This figure is modified from a paper published in 2012 indicating the “pain genes” that had been identified across the genome and what pain types each was associated with. A couple more have been added by now, but fewer than you might think.
Many putative 'pain genes' have indeed been genetically linked to various chronic pain conditions, but study results have proven difficult to replicate and consequently are yet to have real impact on treatment approaches.
Of a wide range of candidates, some have received particular attention from researchers and can be used to illustrate how it is determined that a gene is really a "pain gene". Four examples follow: COMT, encoding an enzyme that eliminates catecholamines (and the somewhat related ADRB2, the beta-adrenergic receptor); GCH1, which encodes GTP cyclohydrolase; and OPRM1, the μ-opioid receptor gene.
Catechol-O-Methyltransferase: The First Pain Gene
Probably the most studied and characterized gene with a role in pain sensitivity and/or susceptibility is COMT. In 2003, it was the first "pain gene" identified.
SNPs within COMT were evaluated for their relationship to both opioid neurotransmitter responses in nucleus accumbens via PET (Positron Emission Tomography) scanning and to reports of pain intensity after hypertonic saline injection.
The authors hypothesized that the low functioning met/met genotype for COMT (i.e., people that were homozygous for the minor allele encoding the methionine substitution at codon 158 (Val158Met)) would be associated with overactive dopamine signalling and less opioid signaling under sustained painful conditions induced by injection of hypertonic saline.
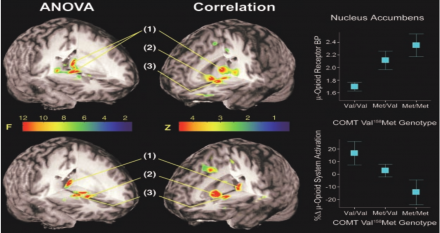
*Image courtesy of Zubieta et al., 2003. Science, 299(5610).
A single SNP in COMT was found to be associated with altered opioid response to painful stimulation AND with higher pain intensity ratings. Subsequently, a lot of work has been done to look at how and why polymorphisms within COMT affect pain. It is one of the best characterized stories because a mechanism of action has been identified.
Interestingly, other genetic variants within the catecholamine neurotransmitter pathways have also been associated with orofacial pain. The beta-2-adrenergic receptor (ADRB2) has also been linked to orofacial pain and fibromyalgia and chronic widespread pain disorder risk and severity, suggesting that it is a gene that plays a role in pain, in general, as opposed to a particular type of pain exclusively.
Comt/COMT and Pain
COMT encodes an enzyme that is important for the breakdown (i.e. inactivation) of catechol substances, including three neurotransmitters (dopamine, norepinephrine and epinephrine).
- Single polymorphism rs4680 results in a Val158Met substitution
- Val/Val homozygotes - activity increases
- Met/Met homozygotes - activity decreases
- 22 frequent polymorphisms in this gene have been analyzed for their correlation to different aspects of pain.
With a Val/Met substitution just at location 158, you have a 3-4 fold change in enzymatic activity that is manifested as lower activity enzyme and slower breakdown of the neurotransmitters in question.
So, higher activity means less neurotransmitter available to work; lower activity means more neurotransmitter.
- Haplotypes (combination of SNPs in a gene)
- High Pain Sensitivity –HPS
- Low Pain Sensitivity – LPS
- Average Pain Sensitivity -- APS
A significant part of the human population develops chronic pain conditions that are characterized by heightened pain sensitivity. Three genetic variants (haplotypes) of the gene encoding catecholamine-O-methyltransferase (COMT) were identified.
These are designated as low pain sensitivity (LPS), average pain sensitivity (APS) and high pain sensitivity (HPS). These haplotypes encompass 96% of the human population, and five combinations of these haplotypes are strongly associated (P=0.0004) with variation in the sensitivity to experimental pain.
The presence of even a single LPS haplotype diminishes, by as much as 2-3 times, the risk of developing myogenous temporomandibular joint disorder (TMD). The LPS haplotype produces much higher levels of COMT enzymatic activity when compared with the APS or HPS haplotypes.
Inhibition of COMT in a rat results in a profound increase in pain sensitivity. Thus, COMT activity substantially influences pain sensitivity, and the three major haplotypes determine COMT activity in humans that inversely correlates with pain sensitivity and the risk of developing TMD.
- Haplotypes (combination of SNPs in a gene)
- High Pain Sensitivity –HPS
- Low Pain Sensitivity – LPS
- Average Pain Sensitivity -- APS
These polymorphisms don’t happen randomly, but rather they tend to occur in groups or haplotypes so that one single SNP can actually tell you with a high likelihood what the genotype is at a block of SNPs in the same region of the gene. In this way, 4 basic blocks of SNPs have been demonstrated to combine into four frequent haplotypes ... and the haplotypes are highly predictive of experimental pain sensitivity.
COMT has been implicated in the sensitivity to experimental pain, migraine, TMD and (based on a small number of studies in rodents) even neuropathic pain. The TMD (temporomandibular disorder) is of particular interest here because COMT significantly impacts pain in this disorder! So, pain sensitivity could be altered by genotype in those showing the most severe pain.
Interestingly, in this case, animal models were used to follow up on the studies to determine “where” the effects were coming from (brain entirely, periphery, or a combination) and to test antagonists that are known to act on this system in the targeted treatment of pain. So, while the original hypothesis was tested in humans, mechanistic investigations were pursued in animal models.
It should also be mentioned that this study was conducted on white (Caucasian) females, but shows a relationship between the high pain sensitivity and risk for the chronic orofacial pain disorder TMD (temporomandibular disorder). Moreover, variation within COMT has been specifically associated with risk of developing TMD and for differential treatment outcomes following diagnosis.
GCH1 is Associated with Pain
The next example is GTP cyclohydrolase (GCH1). GCH1 is a rate limiting step in the formation of tetrahydrobiopterin (BH4) and, when added to the pathway, raises levels of BH4, a molecule that helps enzymes make important neurotransmitters such as serotonin and dopamine.
Researcher Clifford Woolf found that the GCH1 also strongly modulates pain sensitivity.
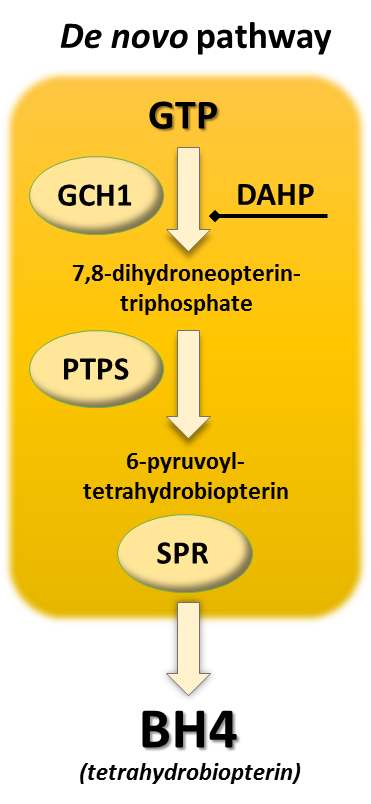
GCH1 is Increased in DRG After Nerve Injury
Pathway analysis found a number of genes related to BH4 production that were elevated in dorsal root ganglian (DRG) after spinal nerve injury (SNI) and GCH1 is a vital metabolic step in that process.
After seeing changes in BH4 and identifying that the difference in expression in the microarray was due to a difference in GCH1 and not the other two enzymatic steps in BH4 biosynthesis, GCH1 became the highest priority candidate gene.
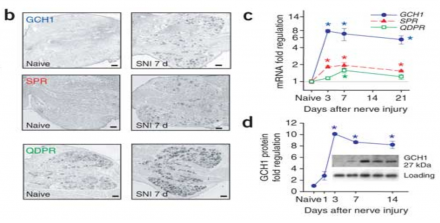
*Images reprinted by permission from Macmillan Publishers Ltd: Nature Medicine Costigan et al. (2006). GTP cyclohydrolase and tetrahydrobiopterin regulate pain sensitivity and persistence. Nature Medicine. (12), 1269-1277
Translational Findings for GCH1
Remember from the chromosome map that GCH1 is found on Chromosome 14, and the vast majority of the subjects tested were homozygous for the more common allele at this location.
The heritability here is marked by codominance, which you can tell, based on the fact that the heterozygotes have an intermediate pain phenotype that is closer to the “average” of the two homozygote phenotypes. This is the same pattern seen in the COMT data.
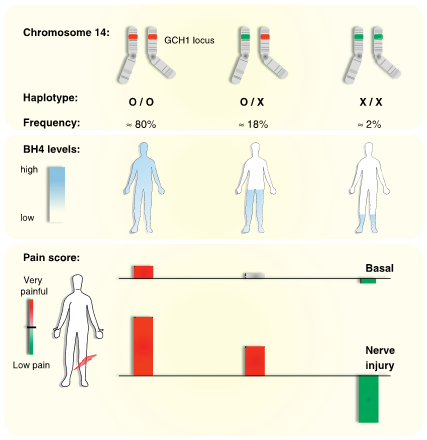
The level of BH4 is directly proportional to allelic identity of the individual with the lowest levels seen in those that are homozygous for the rare allele.
Both Basal and Nerve injury -induced pain scores were significantly predicted by genotype. You can see that the less common genotype results in a reduced pain or analgesic phenotype and that this is even more pronounced following nerve injury.
Therefore, GCH1 not only affects pain sensitivity but also susceptibility to developing chronic pain after nerve injury and in experimental pain paradigms.
If you block synthesis of BH4, pain is reduced. If BH4 is added, it produces pain. BH4 turned out to be implicated in inflammatory pain as well.
As it relates back to the case, if the patient experiences nerve injury as a result of the condition, the neuropathic component could be under differential genetic control, meaning that the short term pain (i.e. acute) could be influenced by a set of genes with variation within them, but that an entirely different (or overlapping but not 100% congruous) set of genes could influence the risk for neuropathic pain.
As neuropathic pain tends to be highly treatment resistant and negatively impacts quality of life over the long term, these genes could prove important as we move toward personalized plans of care that integrate genetic pain risk into the treatment plan.
NIH Context and Role of Researchers
Turning back to humans, Woolf enlisted neurologist Mitchell Max at the NIDCR. Max screened the blood of 168 back-surgery patients for the three pain genes Woolf had investigated in rats to see whether haplotypes of those genes were associated with more or less chronic pain following surgery.
He found that one GCH1 haplotype was highly associated with lower levels of persistent leg pain. Then Max and Roger Fillingim at the University of Florida tested a set of healthy people for the pain-protective GCH1 haplotype and discovered that those with two copies (one from each parent) were less than normally sensitive to thermal, mechanical and deep-muscle pain.
Translational Findings for OPRM1
OPRM1 is the gene encoding the mu opioid receptor, the receptor that is primarily responsible for the analgesic action of opioid drugs like morphine, fentanyl , hydromphone, hydrocodone, methadone, and oxycodone.
The most studied SNP, rs1799971, within OPRM1 (A118G) results in a asparagine to aspartate substitution at position 40 of the protein (N40D). This is a non-synonymous SNP within a coding region of the gene and results in significant differences in function of mu opioid receptor, i.e. how effectively it binds opioid drugs/agonists and how much analgesia patients report as a result of these treatments.
SNPs in the human mu opioid receptor gene alter beta-endorphin binding and activity, and therefore could have possible implications for opiate addiction.
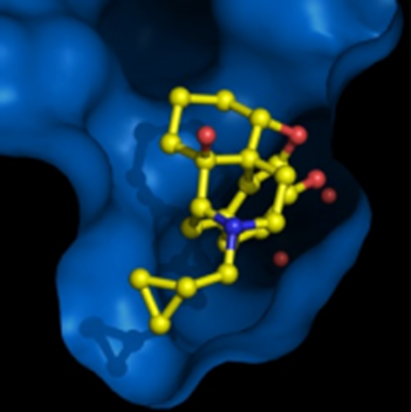
Subjects identified as AA homozygotes, compared with G carriers, (A/G or G/G genotypes, A118G) show greater μ-opioid receptor binding in brain regions that included the anterior cingulate cortex (subgenual, rostral, and dorsal ACC), the ventral striatum (NAc: nucleus accumbens) and the thalamus (THA), all of which play an explicit role in pain perception and modulation.
OPRM1 118G carriers, compared with AA homozygotes, showed an overall reduction of mu receptor binding by morphine in multiple regions implicated in pain, stress, and emotion regulation. Similar patterns of reduced mu opioid receptor binding have also been observed in patients with fibromyalgia, a persistent pain syndrome characterized by chronic widespread pain. Reductions in μ-opioid availability in vivo in limbic regions were associated with greater clinical pain ratings and, in particular, the affective quality of pain.
The key point is that one or two copies of the minor (rare) allele G at this location have higher pain ratings and greater analgesic need because they have less efficient mu-receptor binding. This is linked with endogenous (and exogenous) pain modulation and could be important for not only pain sensitivity but also for pain management and control.
Epigenetics
Epigenetics refers to functionally relevant modifications to the genome that do not involve changes in the nucleotide sequence. Examples of mechanisms that fall under this heading are DNA methylation and histone modification (e.g. acetylation).
These modify how genes are expressed, or how much or how little of the end gene product is made.
When DNA methylation is reduced, gene expression increases.
When histone acetylation increases, gene expression increases.
These processes can be induced within an individual’s lifetime (e.g. by stressful experience, disease, inflammation, diet, exercise, etc.), and the changes can be passed down from generation to generation through the inherited DNA. In this way, the mother’s (or father’s) experience may impact their future childrens’ gene expression, which could be a way for multiple generations to be altered without changes in the DNA sequence.
The impact of the environment would be an example of a stimulus that could induce epigenetic but not genetic (because genotpye is fixed over time) changes.
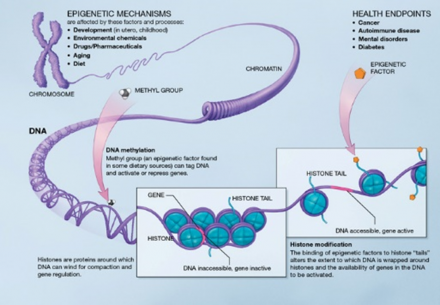
*http://commonfund.nih.gov/epigenomics/figure.aspx from NIH and wikipedia
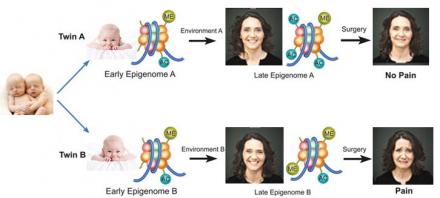
The Epigenome: Setting the Stage for Pain Susceptibility
The epigenome may actually show greater diversity than the genome and that prior experience, history, environment, etc. all impact the epigenome. So much so that identical twins are no more likely to have identical epigenomes than are unrelated strangers.
The methylation, histone acetylation and microRNA content of one’s genome is a highly variable entity. This varies greatly between individuals and is also impacted by transient changes due to experience during ones lifetime.
Life is personal and your epigenome changes accordingly.
Epigenetic Mechanisms
DNA Methylation
- 5-methylation of cytosine nucleotides mediates gene repression
- 5-hydroxymethylation of cytosine produces gene expression
DNA methylation is a process by which methyl groups are added to DNA, thereby modifying DNA function. When located in a region that promotes gene expression, DNA methylation typically acts to suppress gene transcription (so, more methylation, less gene expression).
Hypomethylation typically corresponds to increased gene expression. DNA methylation is essential for normal development, playing a key role in processes including genomic imprinting, X-chromosome inactivation, and aging.
Histones
Acetylation of most histone subunits (at any of several Lysine residues) promotes gene transcription.
Methylation can repress or activate gene transcription:
- H3 methylation of Lys9 or Lys27 associated with gene repression
- H3 methylation of Lys4, Lys36 or Lys79 associated with gene activation
- Phosphorylation, ubiquitination and ADP ribosyltion
A Histone modification is a covalent post-translational modification to histone proteins which includes methylation, phosphorylation, acetylation, ubiquitylation, and sumoylation. The PTMs made to histones can impact gene expression by altering chromatin structure or recruiting histone modifiers. Histone proteins act to package DNA, which wraps around the eight histones, into chromosomes. Histone modifications act in diverse biological processes such as transcriptional activation/inactivation, chromosome packaging, and DNA damage/repair.
miRNA activity
Noncoding functional RNAs that have a critical role in gene transcription
A micro RNA (abbreviated miRNA) is a small non-coding RNA molecule (containing about 22 nucleotides) found in plants, animals, and some viruses, which functions in RNA silencing and post-transcriptional regulation of gene expression.
Recent findings indicate that patients with painful disorders such as CRPS, interstitial cystitis, and IBS have dysregulations in miRNAs that appear to contribute to their pain state. Animal studies reveal a role for miRNAs in pain processing for both inflammatory and neuropathic pain. Within the last six years, accumulating evidence has suggested that these small non-coding inhibitory RNAs known as microRNAs play an important role in regulating pain-processing within a wide range of experimental models and clinical pain disorders.
Family History
If genetics and epigenetics are such powerful influences over pain sensitivity then why isn’t pain sensitivity more random or more familiar? Why do we seem to recognize patterns of pain sensitivity in population groups? Why do we see pain sensitivity patterns that seem to reflect gender, race, ethnicity, or even culture? And why do we see individuals who defy these recognized patterns?
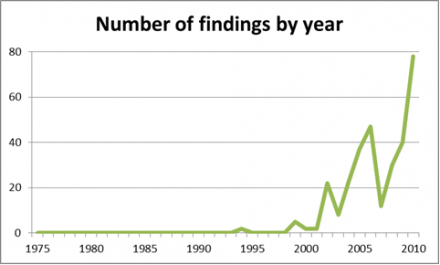
Pain is Polygenic, Multifactorial, and Complex
This graph illustrates the exponential increase in the number of pain-relevant candidate association study findings in humans by year of publication.
It underscores the relative youth of the field and the rapid accumulation of findings in the past 20 years.
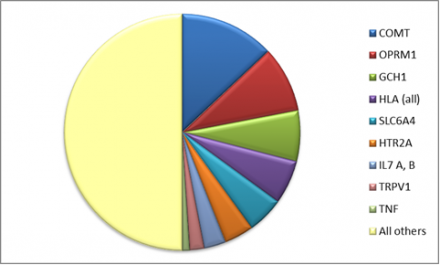
This chart illustrates popular candidate "pain genes”. Publications evaluating the contribution of these nine genes equal studies of all other genes combined.
However, the findings from many of these targets are “mixed”, with replication rates being quite low for key findings between studies.
The pain genes database is a database started and maintained by Jeff Mogil at McGill University, in which all genetic mouse models with a pain phenotype are catalogued.
You can search this database for findings on any gene that has been associated with an altered response to painful stimuli, and a summary of findings and the models used will come up.
http://www.psych.mcgill.ca/pain-apps
Test Your Knowledge
Question 1
The genetic code changes through epigenetics.
Question 2
If the child can be distracted from his pain this usually means that he is not experiencing a high level of pain.
Question 3
Adolescents may sleep in spite of severe pain.
Question 4
Individual variability in pain is most likely due to:
Question 5
The most accurate judge of the intensity of the child's pain is the:

